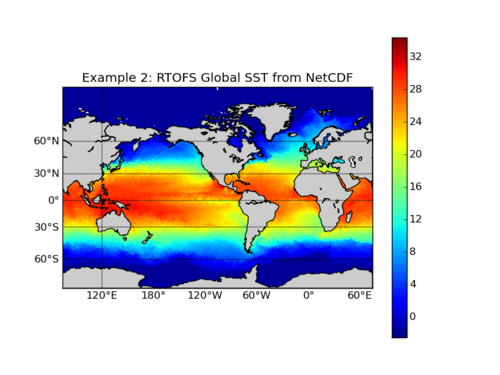
The following example uses Sage Python to extract and visualize the sea surface temperature in the Global RTOFS model using data from the NOMADS data server or a downloaded Global RTOFS NetCDF file.
The examples make use of the following free software:
Begin by importing the necessary modules and set up the figure
from mpl_toolkits.basemap import Basemap import numpy as np import matplotlib.pyplot as plt from pylab import * import netCDF4 plt.figure()
First set up the URL to access the data server.
See the
Global RTOFS directory on NOMADS for the list of
available model run dates.
mydate='20251230'; url='//nomads.ncep.noaa.gov:9090/dods/'+ \ 'rtofs/rtofs_global'+mydate+ \ '/rtofs_glo_3dz_forecast_daily_temp'
Note that the NOMADS data server interpolates and delivers the data on a regular lat/lon field, not the native model grid. To analyze the model output on the native grid you will have to work from a downloaded NetCDF file (see Example 2), or use a NOPDEF url from the Developmental NOMADS. The latter are experimental links that bypass the grid interpolation step. For example, the NOPDEF URL would be as follows:
mydate='20251230'; url='//nomad1.ncep.noaa.gov:9090/dods/'+ \ 'rtofs_global/rtofs.'+mydate+ \ '/rtofs_glo_3dz_forecast_daily_temp_nopdef'
Extract the sea surface temperature field from NOMADS.
file = netCDF4.Dataset(url) lat = file.variables['lat'][:] lon = file.variables['lon'][:] data = file.variables['temperature'][1,1,:,:] file.close()
Plot the field using Basemap. Start with setting the map projection using the limits of the lat/lon data itself:
m=Basemap(projection='mill',lat_ts=10, \ llcrnrlon=lon.min(),urcrnrlon=lon.max(), \ llcrnrlat=lat.min(),urcrnrlat=lat.max(), \ resolution='c')
Lon, Lat = meshgrid(lon,lat) x, y = m(Lon,Lat)
cs = m.pcolormesh(x,y,data,shading='flat', \ cmap=plt.cm.jet)
m.drawcoastlines() m.fillcontinents() m.drawmapboundary() m.drawparallels(np.arange(-90.,120.,30.), \ labels=[1,0,0,0]) m.drawmeridians(np.arange(-180.,180.,60.), \ labels=[0,0,0,1])
colorbar(cs)
plt.title('Example 1: Global RTOFS SST from NOMADS')
plt.show()
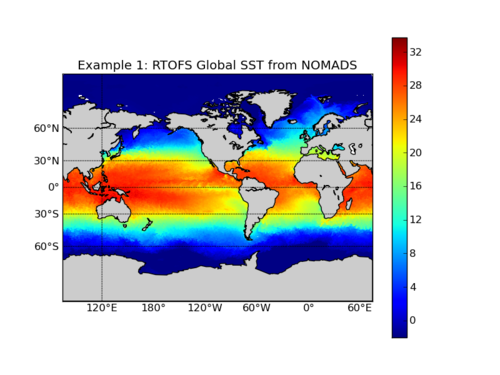
This example requires that you download a NetCDF file from either our NOMADS data server or the Production FTP Server (see our Data Access page for more information. For this exercise, I used the nowcast file for 20111004: rtofs_glo_3dz_n048_daily_3ztio.nc retrieved from NOMADS. This example assumes that the NetCDF file is in the current working directory.
Begin by importing the necessary modules and set up the figure
from mpl_toolkits.basemap import Basemap import numpy as np import matplotlib.pyplot as plt from pylab import * import netCDF4 plt.figure()
nc='rtofs_glo_3dz_n048_daily_3ztio.nc';
file = netCDF4.Dataset(nc) lat = file.variables['Latitude'][:] lon = file.variables['Longitude'][:] data = file.variables['temperature'][0,0,:,:] file.close()
lon = np.where(np.greater_equal(lon,500),np.nan,lon)
Plot the field using Basemap. Start with setting the map projection using the limits of the lat/lon data itself (note that we're using the lonmin and lonmax values computed previously):
m=Basemap(projection='mill',lat_ts=10, \ llcrnrlon=np.nanmin(lon),urcrnrlon=np.nanmax(lon), \ llcrnrlat=lat.min(),urcrnrlat=lat.max(), \ resolution='c')
x, y = m(lon,lat)
cs = m.pcolormesh(x,y,data,shading='flat', \ cmap=plt.cm.jet)
m.drawcoastlines() m.fillcontinents() m.drawmapboundary() m.drawparallels(np.arange(-90.,120.,30.), \ labels=[1,0,0,0]) m.drawmeridians(np.arange(-180.,180.,60.), \ labels=[0,0,0,1])
colorbar(cs)
plt.title('Example 2: Global RTOFS SST from NetCDF')
plt.show()